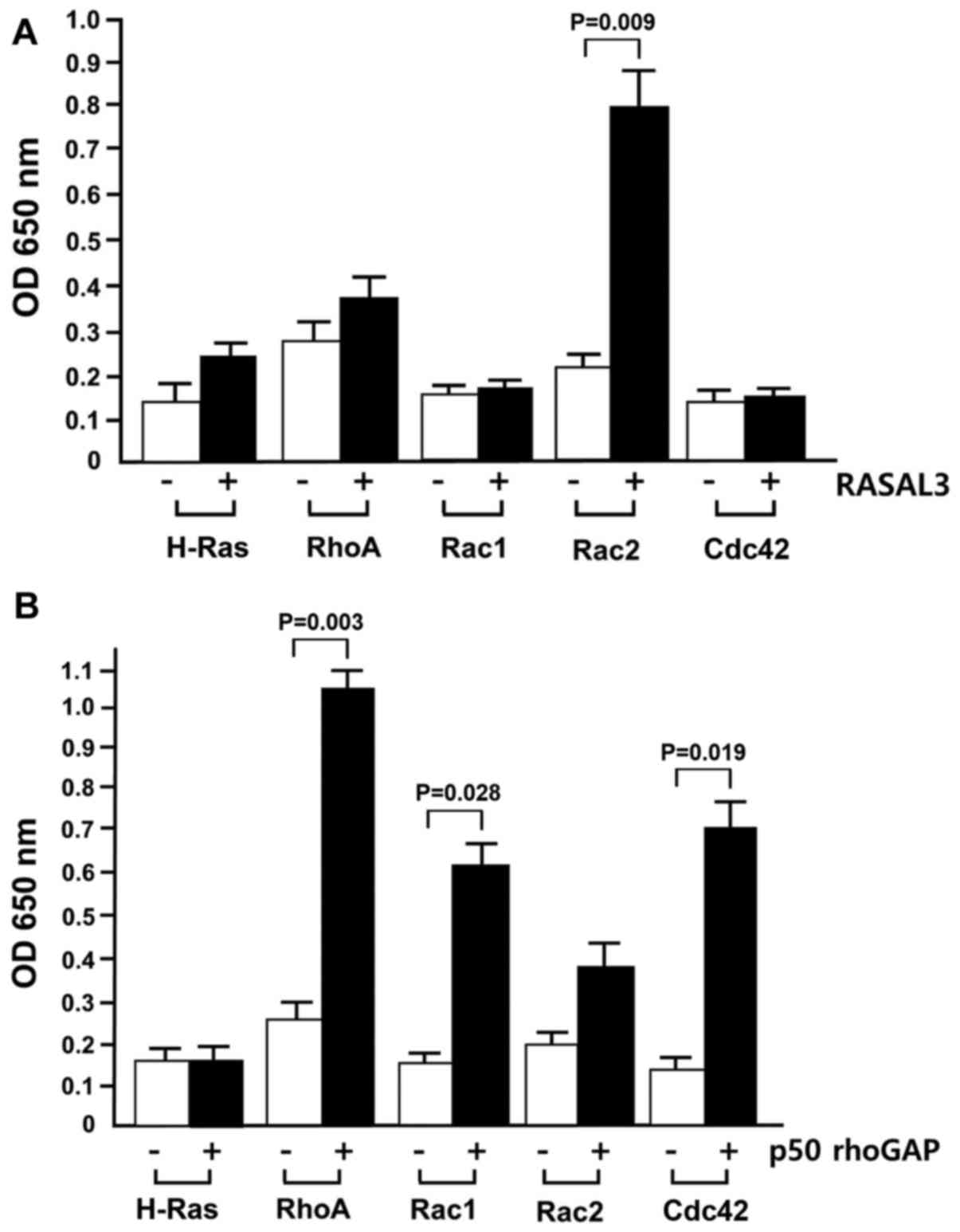
Scheme 1. The QM-derived TS model for GTP hydrolysis by RhoA/RhoGAP... | Download Scientific Diagram

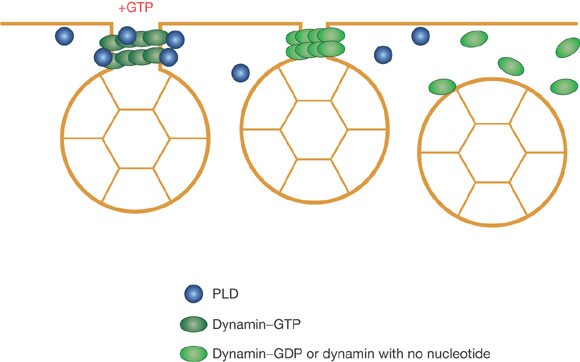
Rab Proteins Regulate Membrane Fusion Through GTP Hydrolysis. (Left)... | Download Scientific Diagram

GTP Hydrolysis Without an Active Site Base: A Unifying Mechanism for Ras and Related GTPases | Journal of the American Chemical Society
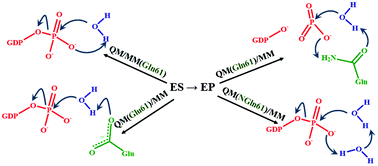
Diversity of mechanisms in Ras–GAP catalysis of guanosine triphosphate hydrolysis revealed by molecular modeling - Organic & Biomolecular Chemistry (RSC Publishing)

Changes in the triphosphate conformation that accelerate hydrolysis by... | Download Scientific Diagram

Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations | PNAS

Stationary points along the wild type Ras.GAP GTP hydrolysis. Breaking... | Download Scientific Diagram

Structural plasticity mediates distinct GAP‐dependent GTP hydrolysis mechanisms in Rab33 and Rab5 - Majumdar - 2017 - The FEBS Journal - Wiley Online Library

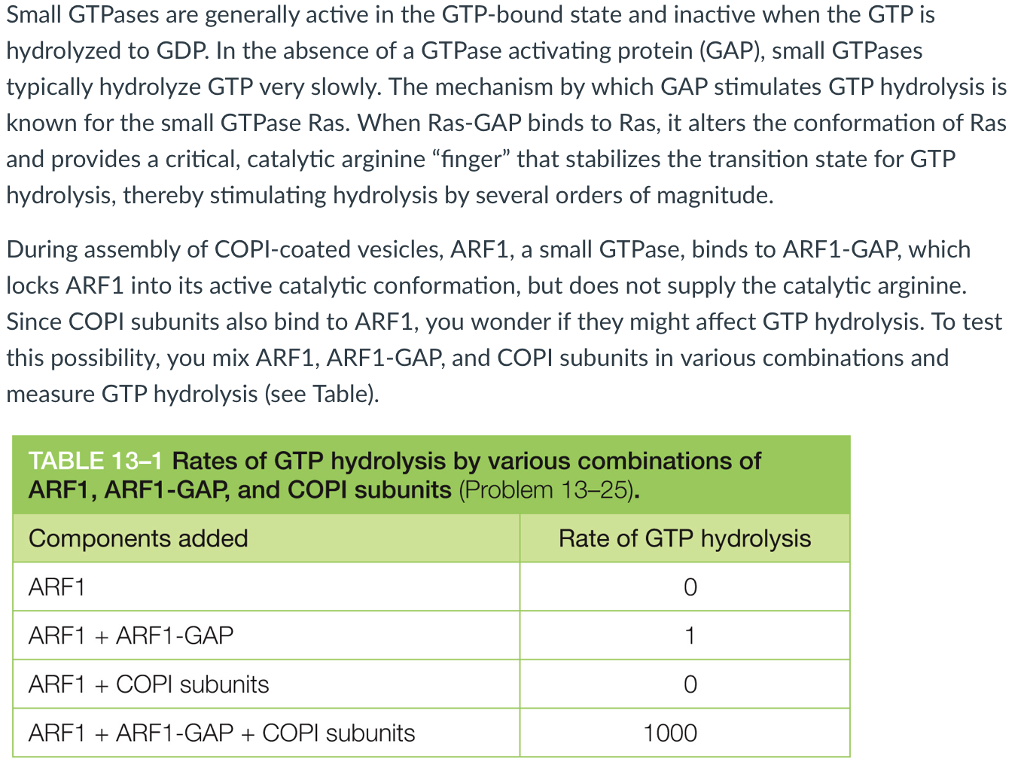
An assay of GAP-catalyzed GTP hydrolysis on Golgi membrane-bound ARF1.... | Download Scientific Diagram

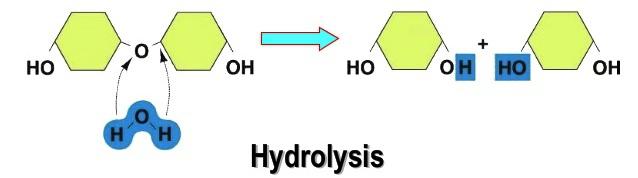
Difference Between Acid Hydrolysis and Enzymatic Hydrolysis | Compare the Difference Between Similar Terms

GAP positions catalytic H-Ras residue Q61 for GTP hydrolysis in molecular dynamics simulations, complicating chemical rescue of Ras deactivation - ScienceDirect
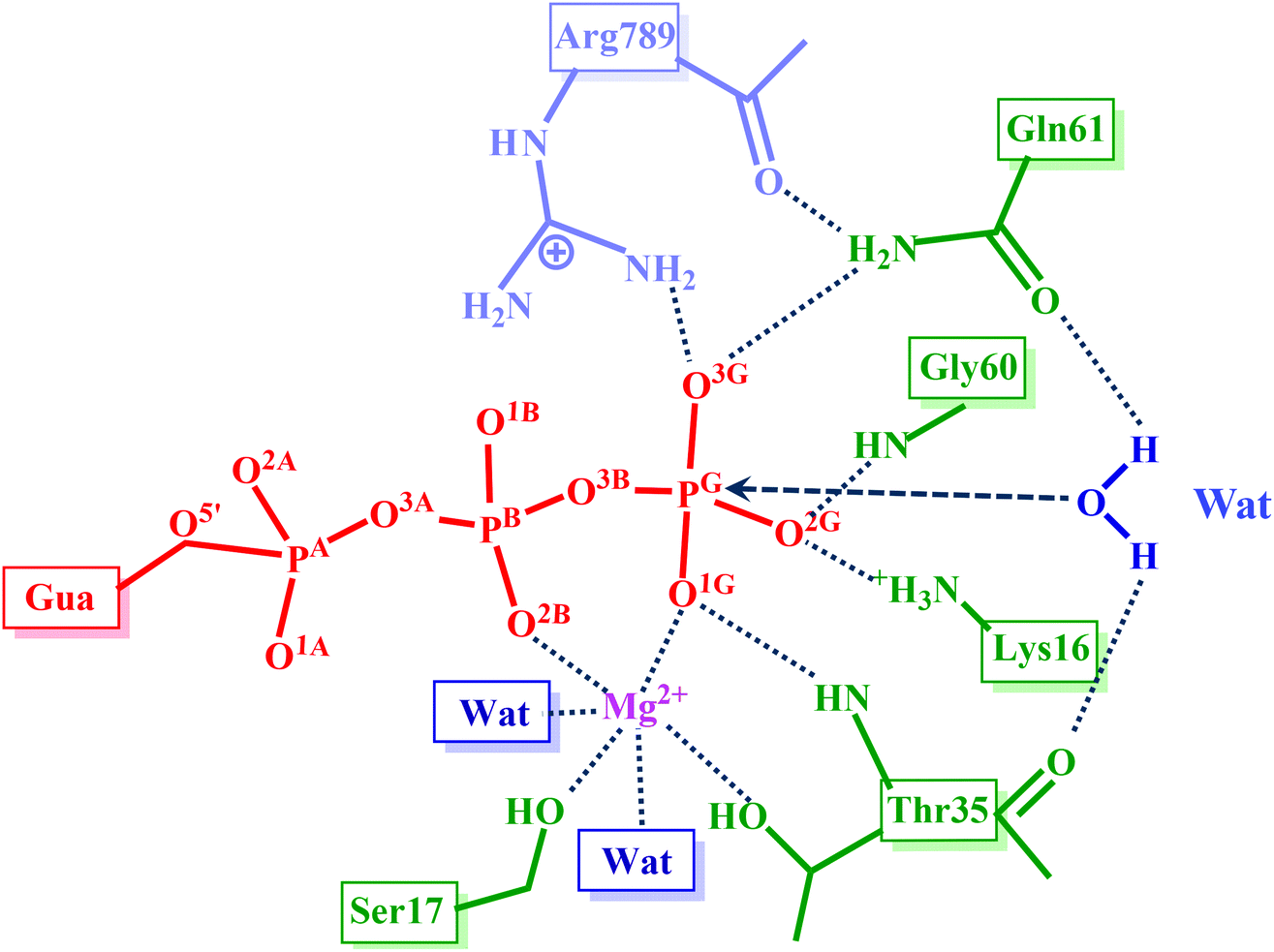
Diversity of mechanisms in Ras–GAP catalysis of guanosine triphosphate hydrolysis revealed by molecular modeling - Organic & Biomolecular Chemistry (RSC Publishing) DOI:10.1039/C9OB00463G

Hydrolysis of Guanosine Triphosphate (GTP) by the Ras·GAP Protein Complex: Reaction Mechanism and Kinetic Scheme | The Journal of Physical Chemistry B

GTP Hydrolysis Without an Active Site Base: A Unifying Mechanism for Ras and Related GTPases | Journal of the American Chemical Society

Phospholipase C-β1 Directly Accelerates GTP Hydrolysis by Gαq and Acceleration Is Inhibited by Gβγ Subunits - ScienceDirect